Our two sister packages are continuously being improved. A brief summary of the latest changes:
shapviz (v0.10.2)
- Identical axes, axis titles and color bars are now collected across dependence plots.
- Dependence plots have received arguments
share_y=FALSEandylim=NULLfor better comparability across subplots. - New visualization for SHAP interaction strenght via
sv_interaction(kind="bar"). It shows mean absolute SHAP interaction/main effects, where the interaction values are multiplied by two for symmetry.
kernelshap (v0.9.1)
permshap()now offers a balanced sampling version which iterates until convergence and returns standard errors. It is used by default when the model has more than eight features, or by settingexact=FALSE.- Fixed an error in
kernelshap()which made the resulting values slightly off for models with interactions of order three or higher. Now, the exact version returns the same values as exact permutation SHAP and agrees with the exact explainer in Python’s shap package.
Illustrating sampling permutation SHAP
Let’s use a beautiful dataset on medical costs to fit a log-linear Gamma GLM with interactions between all features and smoking, and explain it by SHAP on log prediction (= linear) scale.
Since the model does not contain interactions of order above 2, the SHAP values perfectly reconstruct the estimated model coefficients, see our recent paper on https://arxiv.org/abs/2508.12947 for a proof.
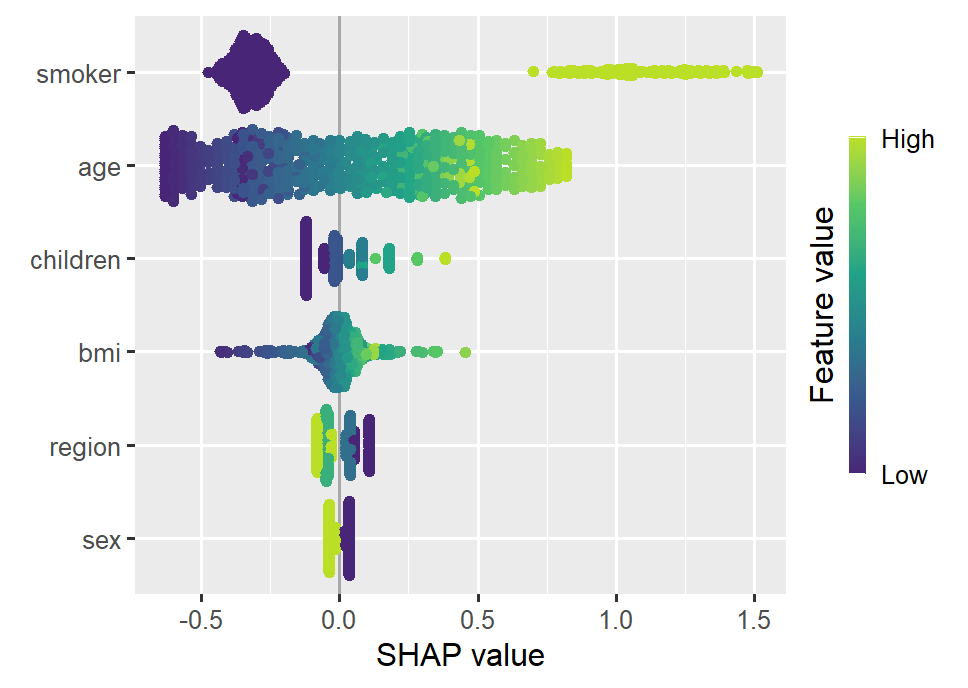
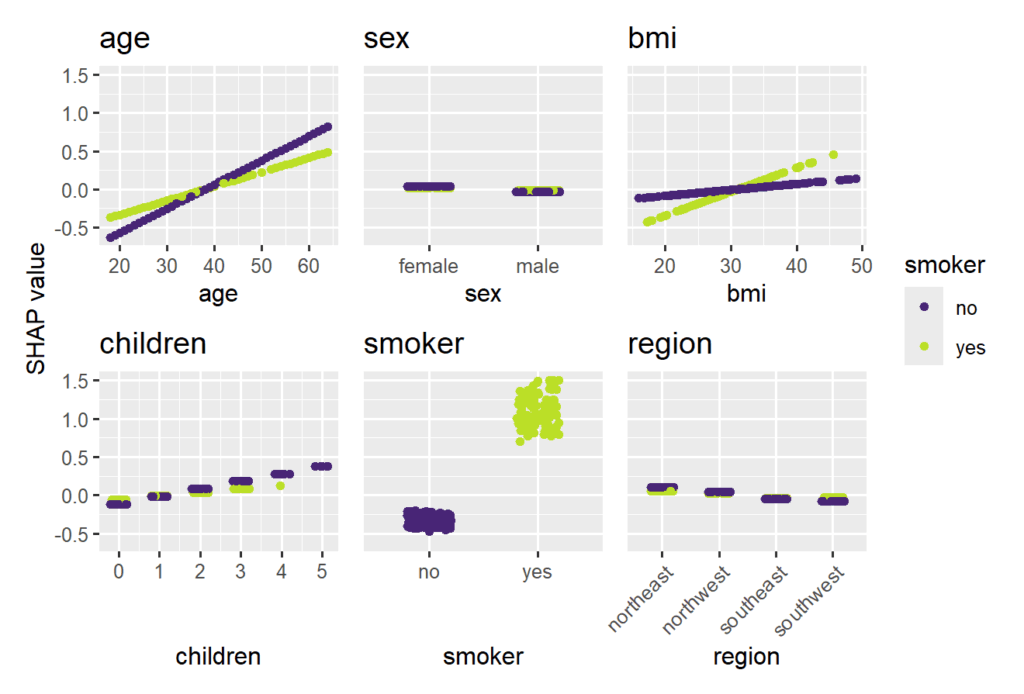
Smoking and age are the most important features. Some strong interactions with smoking are visible.
library(xgboost)
library(ggplot2)
library(patchwork)
library(shapviz)
library(kernelshap)
options(shapviz.viridis_args = list(option = "D", begin = 0.1, end = 0.9))
set.seed(1)
# https://github.com/stedy/Machine-Learning-with-R-datasets
df <- read.csv("https://raw.githubusercontent.com/stedy/Machine-Learning-with-R-datasets/refs/heads/master/insurance.csv")
# Gamma GLM with interactions
fit_glm <- glm(charges ~ . * smoker, data = df, family = Gamma(link = "log"))
# Use SHAP to explain
xvars <- c("age", "sex", "bmi", "children", "smoker", "region")
X_explain <- head(df[xvars], 500)
# The new sampling permutation algo (forced with exact = FALSE)
shap_glm <- permshap(fit_glm, X_explain, exact = FALSE, seed = 1) |>
shapviz()
sv_importance(shap_glm, kind = "bee")
sv_dependence(
shap_glm,
v = xvars,
share_y = TRUE,
color_var = "smoker"
)
Illustrating SHAP interaction strength
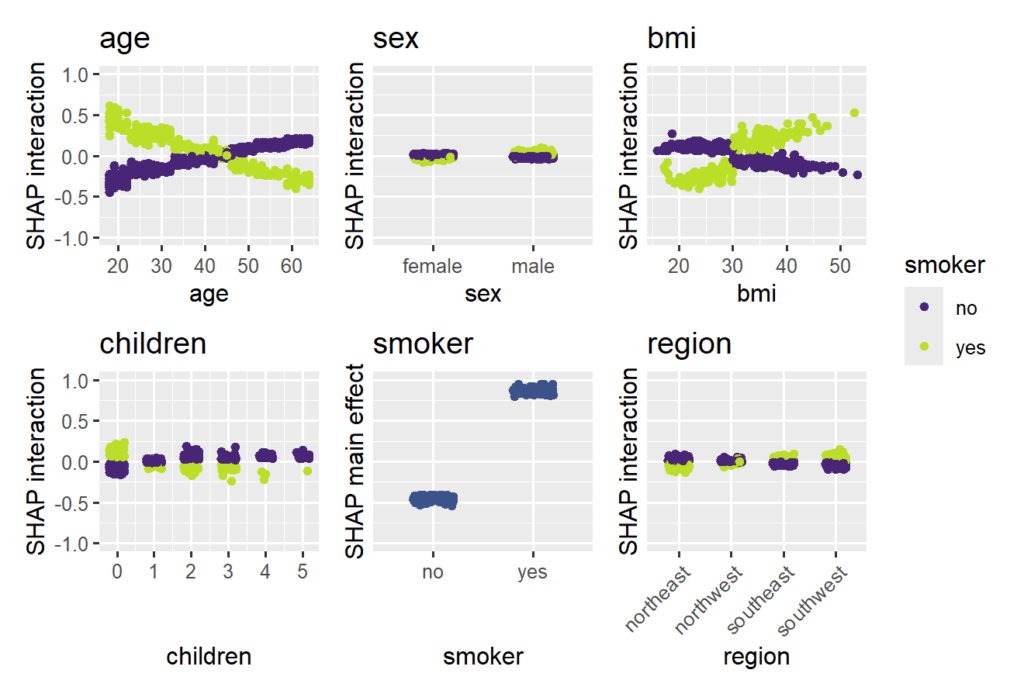
As a second example, we sloppily fit an XGBoost model with Gamma deviance loss to illustrate some of the SHAP interaction functionality of {shapviz}. As with the GLM, the SHAP values are being calculated on log scale.
For the sake of brevity, we focus on plots visualizing SHAP interactions. In practice, make sure to use a clean train/test/(x-)validation and tuning approach.
The strongest interaction effects (smoker * age, smoker * bmi) are stronger than most main effects. Not all interactions seem natural.
# XGBoost model (sloppily without tuning) X_num <- data.matrix(df[xvars]) fit_xgb <- xgb.train( params = list(objective = "reg:gamma", learning_rate = 0.2), data = xgb.DMatrix(X_num, label = df$charges), nrounds = 100 ) shap_xgb <- shapviz(fit_xgb, X_pred = X_num, X = df, interactions = TRUE) # SHAP interaction/main-effect strength sv_interaction(shap_xgb, kind = "bar", fill = "darkred") # Study interaction/main-effects of "smoking" sv_dependence( shap_xgb, v = xvars, color_var = "smoker", ylim = c(-1, 1), interactions = TRUE ) + # we rotate axis labels of *last* plot, otherwise use & guides(x = guide_axis(angle = 45))

Keep an eye on these two packages for further improvements… 🙂